Easy read on Next Generation Sequencing in 3 Mins
Learn about Next Generation Sequencing
Next generation sequencing (NGS), hugely parallel or deep sequencing are related terms that describe a DNA sequencing technology that has revolutionised genomic research. Using NGS, a complete human genome can be sequenced within a single day.
In contrast, the former Sanger sequencing technology used to decode the human genome, required over a decade to deliver the final draft. Although in genome research NGS has mostly supplanted conventional Sanger sequencing, it has not yet translated into routine clinical practice
There are several different NGS platforms using diverse sequencing technologies, a detailed discussion of which is beyond the scope of this discussion. Nevertheless, all NGS platforms execute sequencing of millions of small fragments of DNA in parallel. Bioinformatics analyses are used to fix together these fragments by mapping the individual reads to the human reference genome.
NGS can be employed to sequence complete genomes or confined to specific areas of interest, including all 22 000 coding genes- a whole-exome or small numbers of individual genes.
Next Generation Sequencing systems, introduced in the past decade that allow for massively parallel sequencing reactions. These systems are capable
Sample Preparation
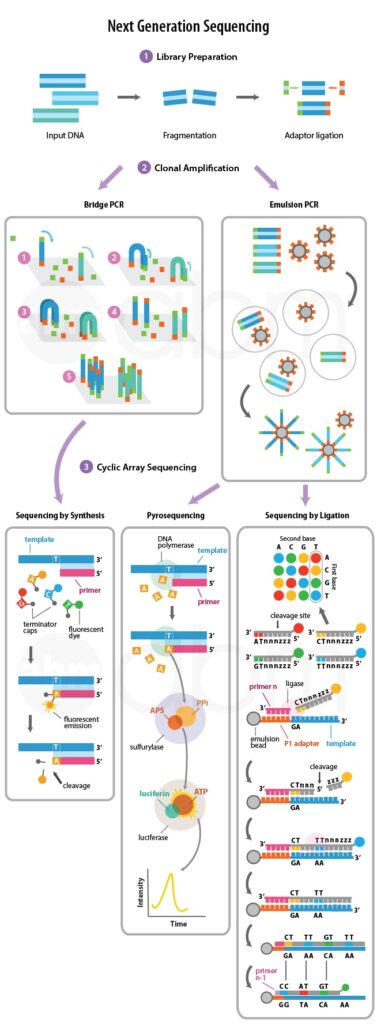
All Next Generation Sequencing platforms entail a library obtained either by amplification or ligation with custom adapter sequences. These adapter sequences allow for library hybridisation to the sequencing chips and provide a universal priming site for sequencing primers. Learn more about sample preparation from our Next Generation Sequencing – Experimental Design knowledge base.
Sequencing machines
Each library fragment, amplified on a solid surface – either beads or flat silicon derived surface, with covalently attached DNA linkers that hybridise the library adapters. This amplification generates clusters of DNA, each originating from a single library fragment; each cluster will act as an individual sequencing reaction.
The sequence of each cluster is optically read (either through the generation of light or fluorescent signal) from repeated cycles of nucleotide incorporation. Each machine has its unique cycling condition – for example, the Illumina system uses repeated cycles of incorporation of reversibly fluorescent and terminated nucleotides followed by signal acquisition and removal of the fluorescent and terminator groups.
Data output
Each machine offers raw data at the end of the sequencing run. This raw data is a group of DNA sequences, generated at each cluster. This data could be further analysed to provide more meaningful results.
The contrasts between the different Next Generation Sequencing platforms lie mainly in the technical details of the sequencing reaction. Below we describe these technical differences briefly. For a full explanation, please visit the manufacturers’ webpages at the links provided in each section.
Pyrosequencing
In pyrosequencing, the sequencing reaction monitored through the release of the pyrophosphate during nucleotide incorporation. A single nucleotide added to the sequencing chip, which will lead to its incorporation in a template-dependent manner.
This incorporation will result in the release of pyrophosphate, which is used in a series of chemical reactions ensuing in the generation of light. Light emission is sensed by a camera which records the appropriate sequence of the cluster. Any unincorporated bases are degraded by apyrase before the addition of the next nucleotide.
Demerits
High reagent cost
High error percentage over strings of 6 or more single-base nucleotides.
Sequencing by Synthesis
Sequencing by synthesis utilises the step-by-step incorporation of reversibly fluorescent and terminated nucleotides for DNA sequencing. The Illumina NGS platform uses it. The nucleotides used in this method have been modified in two ways:
- Each nucleotide is reversibly attached to a single fluorescent molecule with unique emission wavelengths.
- Each nucleotide is also reversibly terminated, ensuring that only a single nucleotide will be incorporated per cycle.
All four nucleotides are appended to the sequencing chip, and after nucleotide incorporation, the remaining DNA bases are washed away. The fluorescent signal is inspected at each cluster and documented; both the fluorescent molecule and the terminator class are then cleaved and washed away. This procedure is reiterated until the sequencing reaction is complete.
Demerits
As the sequencing reaction continues, the error rate of the machine also increases, and it is due to partial removal of the fluorescent signal, which leads to higher background noise levels.
Sequencing by Ligation
Sequencing by ligation is different from the other two methods since it does not utilise a DNA polymerase to incorporate nucleotides. Instead, it relies on short oligonucleotide probes ligated to one another. These oligonucleotides consist of 8 bases (from 3’-5’): two probe specific bases – there are a total of 16 8-mer probes which all differ at these two base positions and six degenerate bases; one of four fluorescent dyes attached at the 5’ end of the probe.
The sequencing reaction commences by binding of the primer to the adapter sequence and then hybridisation of the appropriate probe. The two probe specific bases guide this hybridisation of the probe and upon annealing, is ligated to the primer sequence through a DNA ligase. Unbound oligonucleotides, washed away, the signal is detected and recorded, the fluorescent signal is cleaved (the last three bases), and then the next cycle commences. After approximately seven cycles of ligation the DNA strand is denatured and another sequencing primer, offset by one base from the previous primer, is used to repeat these steps – in total five sequencing primers are used.
Demerits
This method leads to very short sequencing reads.
Ion Semiconductor Sequencing
Ion semiconductor sequencing utilises the release of hydrogen ions during the sequencing reaction to detect the sequence of a cluster. Each cluster is located directly above a semiconductor transistor which is capable of detecting changes in the pH of the solution. During nucleotide incorporation, a single H+ released into the solution, and the semiconductor detects it. The sequencing reaction itself proceeds similarly to pyrosequencing but at a fraction of the cost.
Demerits
High error rate over homopolymer.
If you want to learn Bioinformatics as coursework with a certificate. Check our Bioinformatics course from Ampersand Academy. Also read about Biostatistics.